Single-Molecule Movies of the Interplay between DNA Polymerase and Single Strand DNA Binding (SSB) Protein
An example conference paper
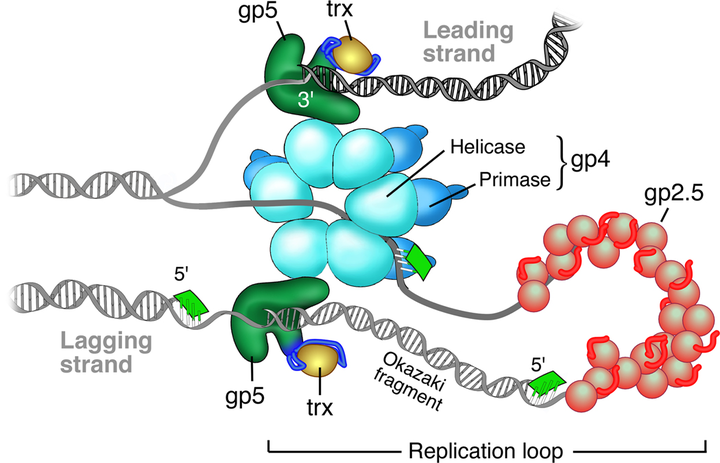 Model of the T7 replisome.
Model of the T7 replisome.Abstract
Transitionally exposed ssDNA during replication are coated and stabilized by single-stranded DNA binding protein (SSB) within the replisome. How these SSB protein regulates and are displaced by DNA polymerase (DNAp) however are still poorly understood. Here T7 DNAp and T7 gp2.5 (SSB) are used as model proteins to investigate their interaction. Single-molecule force spectroscopy study showed that T7 gp2.5 affects DNA replication rate in a force-dependent manner: slowing down DNAp rate at 20pN when fewer secondary structure forming on ssDNA while accelerating replication at 10pN when ssDNA containing possible hairpins, suggesting gp2.5 regulates replication by controlling secondary structure formation. Further dual-color imaging of the dynamics of two molecules revealed that gp2.5 remains stationary while DNAp moves toward gp2.5 to synthesized DNA, supporting the hypothesis of gp2.5 been removed one-by-one rather than pushing all the gp2.5 filament. To gain insight into the mechanism of gp2.5 displacement, we compared unbinding dynamics between wide-type gp2.5 and mutant gp2.5 under various tension, and revealed that wide-type gp2.5 with a c-terminal tail dissociate quicker than that without c-terminal tail, suggesting a molecular role of c-terminal tail could promote quicker unbinding from ssDNA rather than diffusion on ssDNA. Taken together, these data suggest that SSB plays its role by reducing DNA secondary structure and that DNAp removes gp2.5 in a one-by-one manner.